The full EMD 1.0 spec is summarized below; details follow.
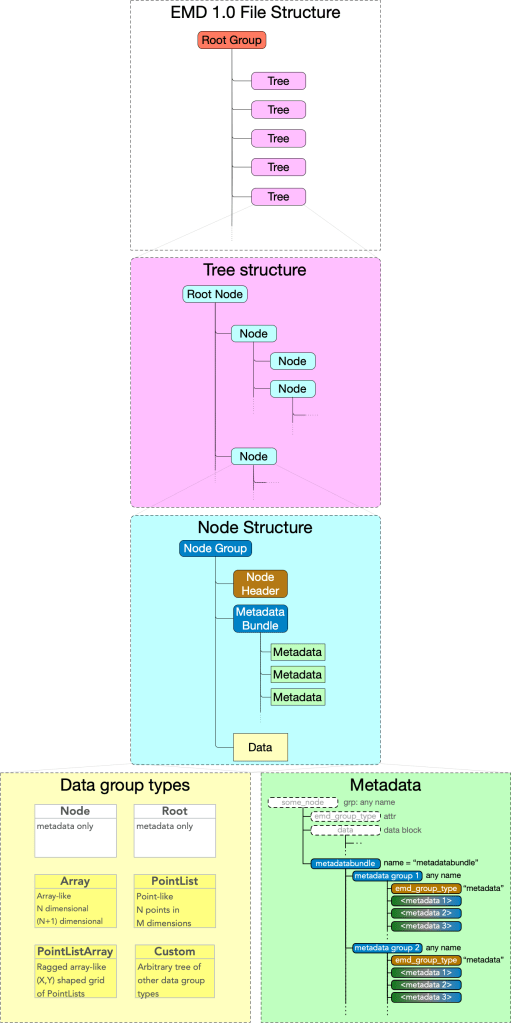
- An EMD 1.0 file contains a root group containing any number of EMD trees.
- An EMD tree is directory-like tree with a root node and any number of nodes.
- A node may contain one data group and any number of metadata groups.
Data groups contain one block of data, plus some self-descriptive metadata. The structure of that data and metadata is determined by the data group type.
Metadata groups contain any number of string named blocks of data of a variety of types, nested to any depth.
In the sections that follow, we detail how each of these abstractions – the EMD 1.0 root groups, trees, (root) nodes, data groups, and metadata groups – map to concrete HDF5 components.
Specification Vs. Format – The HDF5 Format – HDF5 Components
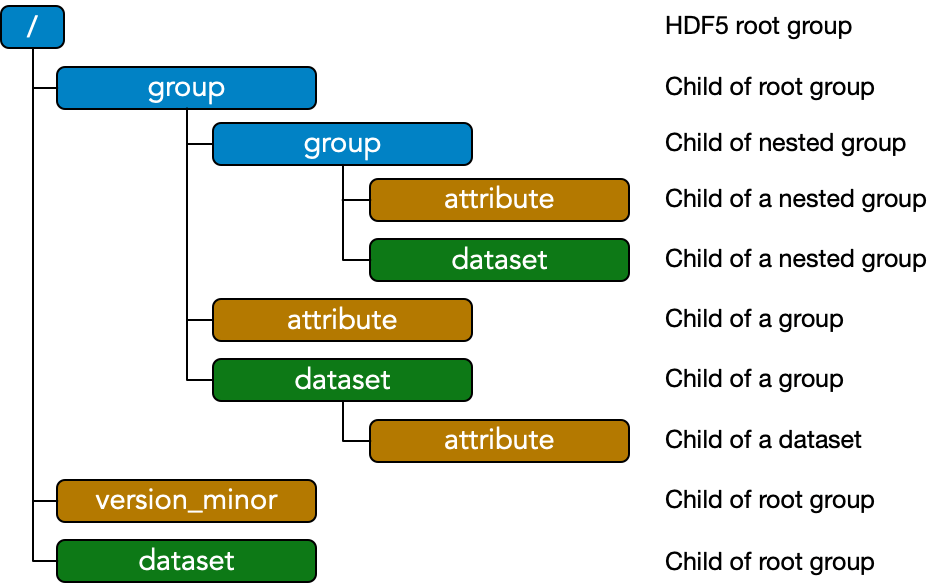
The EMD 1.0 specification is implementation agnostic: files may be built to the EMD 1.0 standard using any sufficiently flexible low-level filetype. The standard EMD 1.0 format is an implementation in the HDF5 file format. Descriptions and schematics here are given in terms of HDF5 components for concreteness using the color scheme above.
The EMD 0.1 Spec
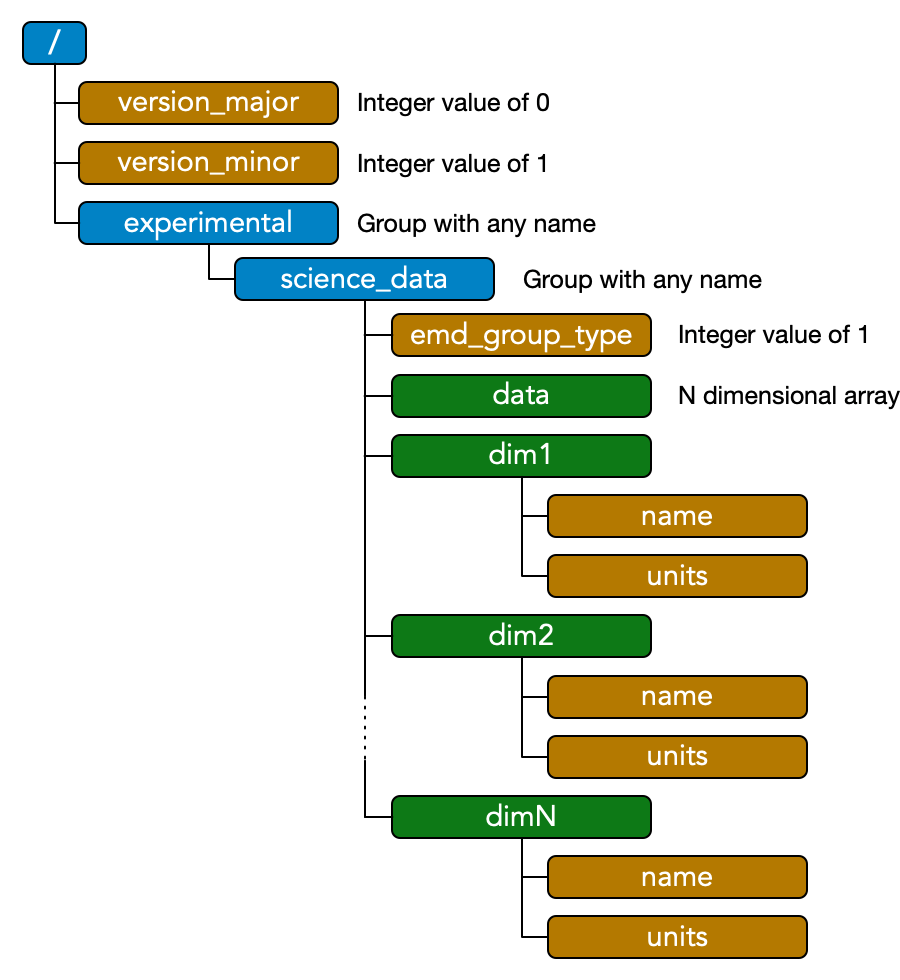
The EMD 1.0 specification builds on and extends the 2012 v0.1 spec. Array-type EMD 1.0 nodes are essentially identical to EMD 0.1 data groups. The schematic shown right represents a minimal EMD 0.1 file, summarizing the spec requirements which can also be found in full here.
Root groups
The EMD 1.0 root group is the file root, concretely in HDF5 the root group, and must contain a specific set of attributes collectively referred to as the file header. There are three required attributes and three optional attributes. The required attributes are
- attr: “emd_group_type” = “file”
- attr: “version_major” = 1
- attr: “version_minor” = 0
where ‘attr:’ specifies an HDF5 attribute, the name of which is given next followed by it’s value to the right. The optional attributes are
- attr: “UUID”
- attr: “authoring_user”
- attr: “authoring_program”
Under the root group there may be any number of EMD trees.

Trees
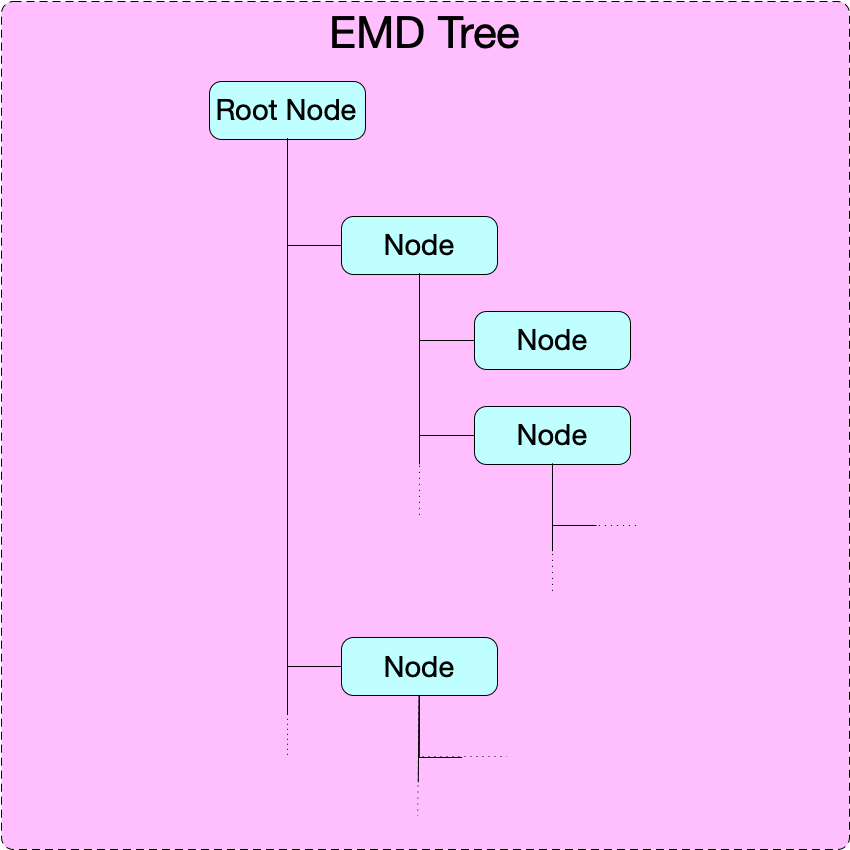
An EMD 1.0 tree is a directory-like tree: it consists of a root node and a set of downstream nodes nested to any depth. A minimal EMD tree requires only a root node. No specification or requirements are made for links and linking.
Nodes
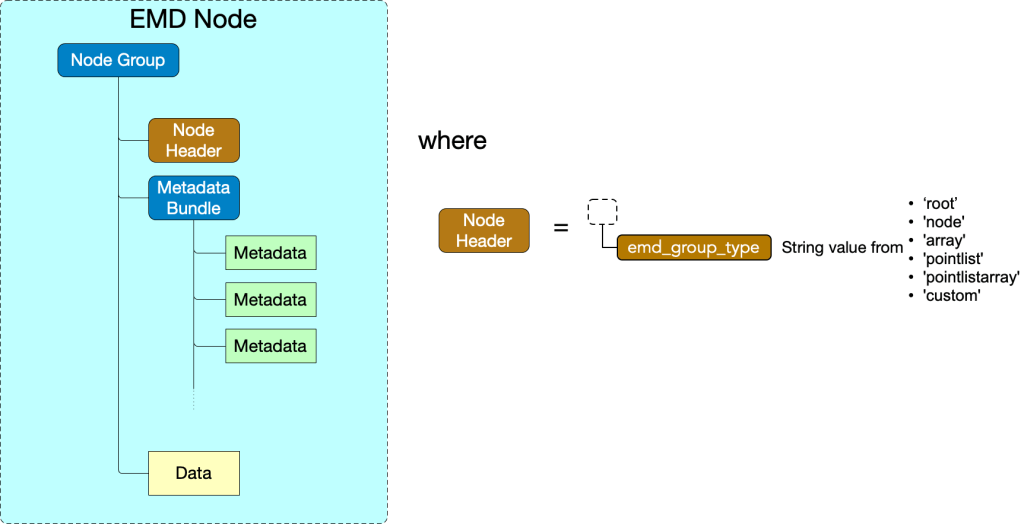
An EMD 1.0 node is an HDF5 group with any name which must have an
- attr: “emd_group_type”
and which may optionally contain
- an EMD data group, consisting of attributes, datasets, and groups defined by the specifications below according to the node’s group type, specified by the “emd_group_type” value
- any number of HDF5 groups corresponding to EMD nodes
- an HDF5 group named “metadatabundle” which contains any number of HDF5 groups corresponding to EMD metadata groups
The generic EMD node structure is shown below.
Data group Types
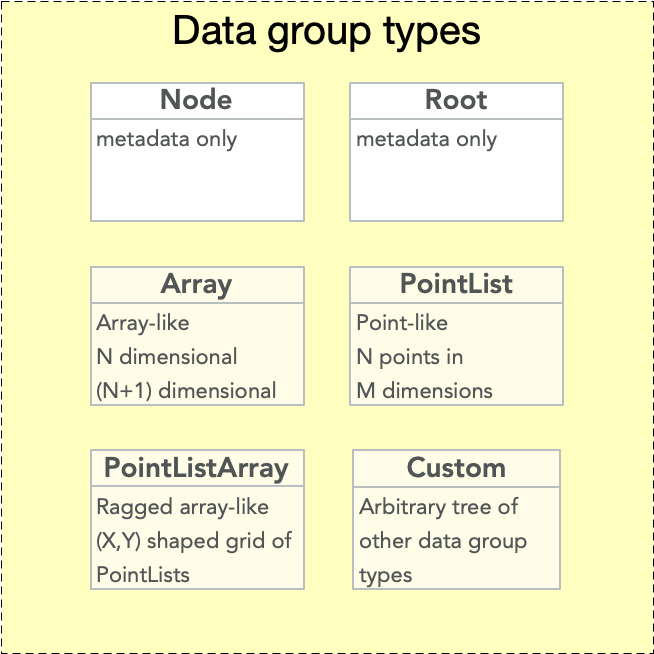
EMD 1.0 nodes may be any one of several possible types. The node type specifies the structure of it’s data group. “Node type” or “data group type” or “data node type” may therefore be used interchangeably.
The node type is specified by the “emd_group_type” attribute. Valid values are “root”, “node”, “array”, “pointlist”, “pointlistarray”, and “custom”. Note that two group types, root and node, contain no data group. The remaining types – array, pointlist, pointlistarray, and custom – must contain data groups.
Group Type: Root node
An EMD 1.0 root node is an HDF5 group nested directly underneath the file root group with any name which must have an
- attr: “emd_group_type” = “root”
and which optionally may contain
- any number of HDF5 groups corresponding to EMD nodes
- an HDF5 group named “metadatabundle” which contains any number of HDF5 groups corresponding to EMD metadata groups.

Root nodes must always be directly underneath the file root group. Each root node is base of a single EMD tree. Every EMD tree must begin with a root node.
Note that EMD 1.0 root group refers to the EMD file root, and EMD 1.0 root nodes refers to EMD tree roots. Root nodes are always one level below the root group. If the file root group and tree root nodes need to be disambiguated explicitly, “file root group” or “HDF5 root group” or “header group” can be used for the EMD root group, and “tree root” or “tree root node” or “tree root group” to refer to the tree root nodes.
Group Type: Node
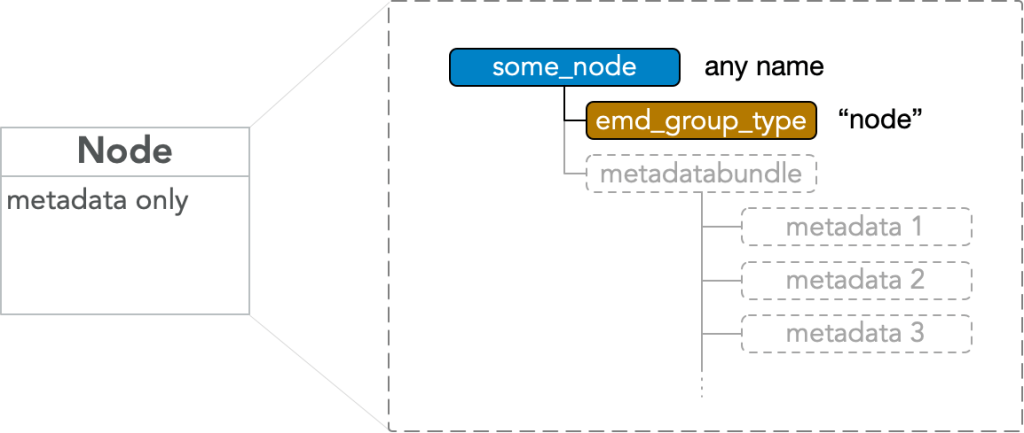
An EMD 1.0 node group is an HDF5 group with any name which must have
- attr: “emd_group_type” = “node”
and which optionally may contain
- any number of HDF5 groups corresponding to EMD nodes
- an HDF5 group named “metadatabundle” which contains any number of HDF5 groups corresponding to EMD metadata groups.
Note that “node” may refer to either the generic structure, where “emd_group_type” is any of the valid types, or to the specific structure above, where the “emd_group_type” value is “node” which has no data block (though may contain metadata). If the distinction needs to be made and is not clear from context, an EMD node of the “node” data group type may be referred to as a “node type group” or “bare node”, and a generic node may be referred to as an “EMD node” or “tree node” or “generic node group”.
Group Type: Array
An array node contains an N dimensional array-like block of data. It must have
- attr: “emd_group_type” = “array”
- dset: “data” = an N dimensional array
where dset: “data” is a dataset with an attr: “units” equal to some string value. The node further contains N additional datasets
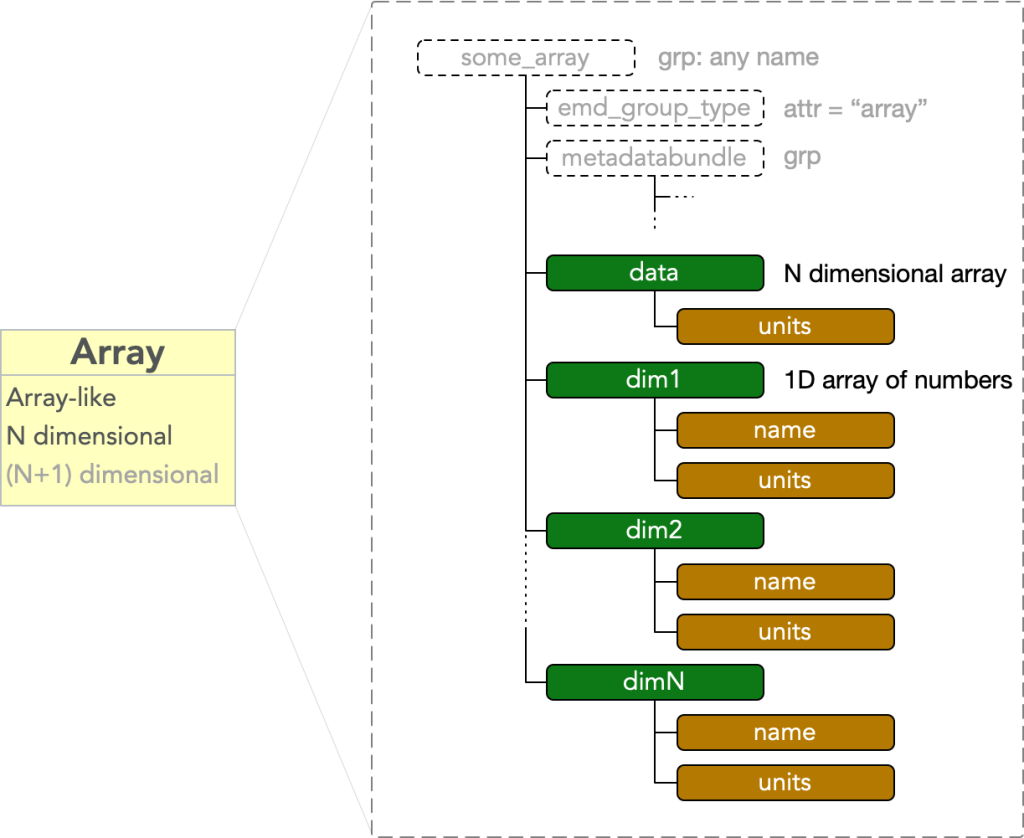
- dset: “dim#”
for each # in (1, …, N), where the data stored in each of these dsets are 1D vectors which calibrate the N axes of the “data” dset. These each have attributes
- attr: “units”
- attr: “name”
attached to each dim# dataset. An array node is therefore a bare EMD node plus a data block consisting of N+1 HDF5 datasets: one called “data”, and N called “dim1” through “dimN”.
Dim vectors
The N dimensional calibration vectors calibrate the primary data array. The i’th vector’s length should match the length of the i’th data array dimension and it’s values specify the coordinates of the corresponding pixel positions in the data array. Dim vectors may be linear or non-linear. If a dimension is linear, i.e. the pixel step size is constant, the length requirement may optionally be relaxed: in this case, a length-2 vector may be stored instead, corresponding to the coordinate positions of the first and second pixels. For example, the x dimension of a 1024^2 pixel micrograph with a pixel size of 0.02 nm would have dim1 values of [0, 0.02, 0.04, … 20.46], and the dataset component of this dim vector may store either this 1D array or the length-2 array [0, 0.02].
Array variant: stack arrays
In addition to the array node specification above, EMD identifies a variant intended to represent an N+1 dimensional array consisting of a stack of N dimensional arrays of any depth L, all sharing a single coordinate system.
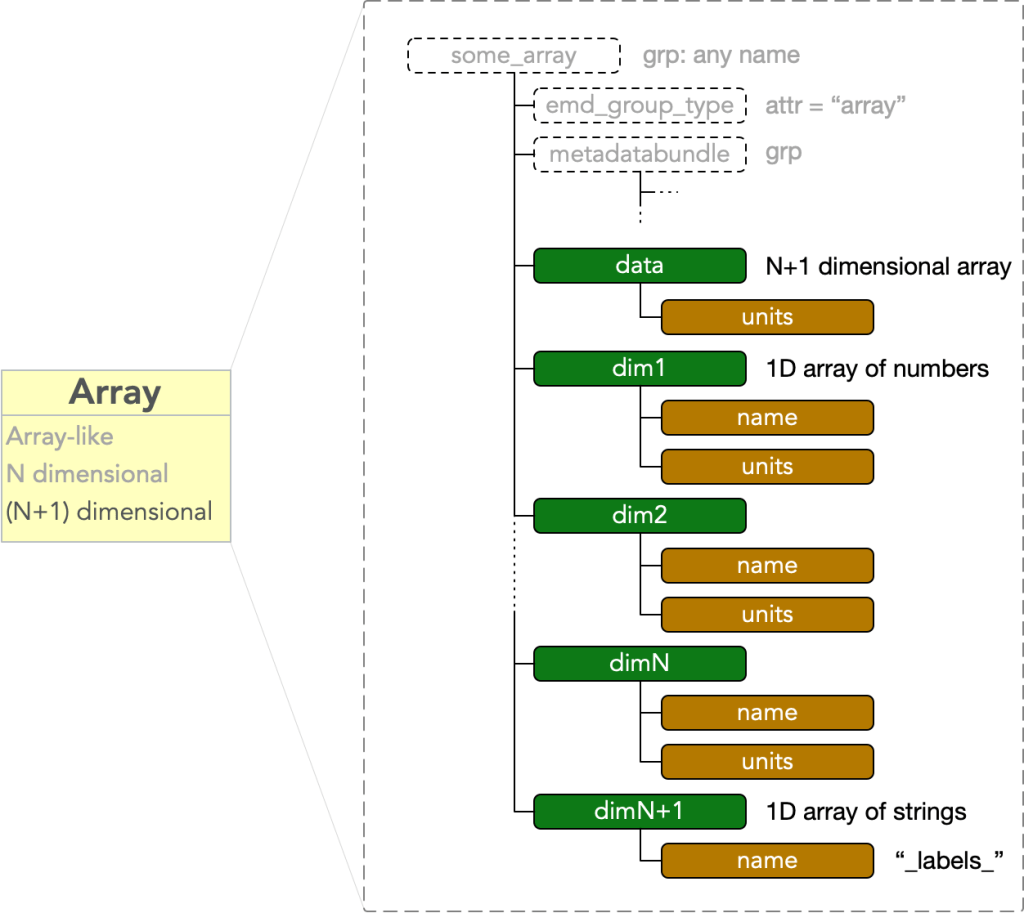
In a stack array, the final dim vector is a length L array of UTF-8 encoded strings rather than numbers, which label the stack of N-D arrays . It must have
- attr: “name” = “_labels_”
and does not have a “units” attribute.
The remaining “dim1” through “dimN” dimension vectors are as described in the standard array node variant, and calibrate each of the N-D arrays in the stack.
Group Type: PointList
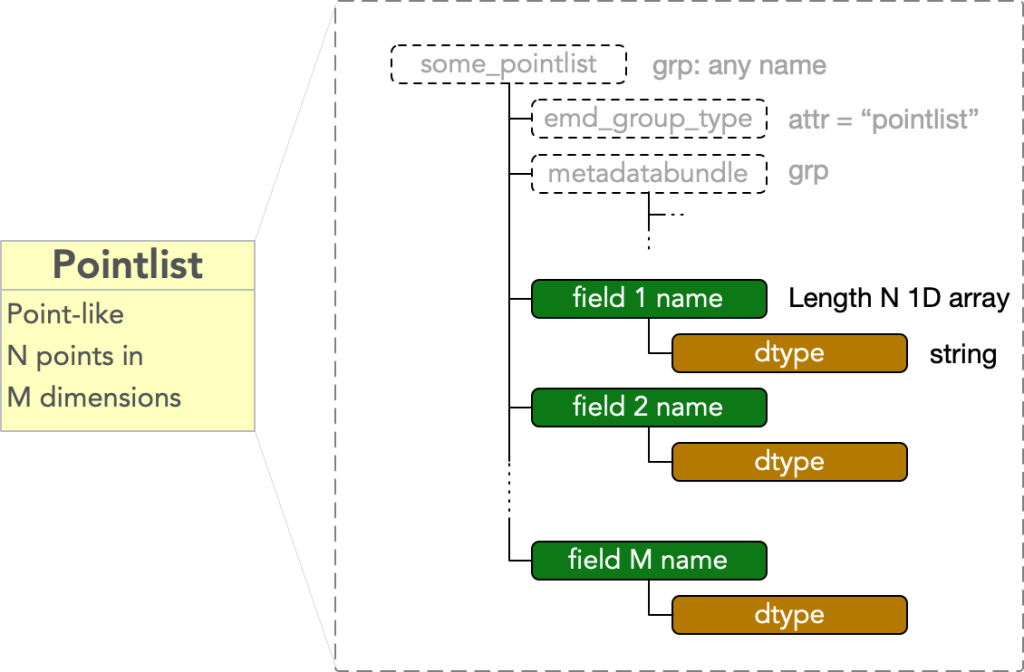
A pointlist type node contains data representing N points in an M dimensional space. It must have
- attr: “emd_group_type” = “pointlist”
Call each of the M dimensions comprising the space in which the pointlists data is embedded a field. Then there are M fields, and each consists of a
- dset: any name = 1D array of length N
and the dataset must have an
- attr: “dtype”
- attr: “units”
equal to a UTF-8 string specifying a valid numpy datatype. The fields in a pointlist are therefore unordered, and the values in each may be represented in different datatypes.
Group Type: PointListArray
A pointlistarray type node holds data structured in an N-D grid, with pointlist-like data at each grid point. The pointlists share a single set of fields, i.e. are all embedded in the same M dimensional space, however, the lengths of the pointlists at each grid point are independent. In the simplest case there is one field (i.e. M=1), and the data may be thought of as an N+1 dimensional ragged array, so called for it’s uniformity and non-uniformity, respectively, along it’s first N dimensions and last dimension.
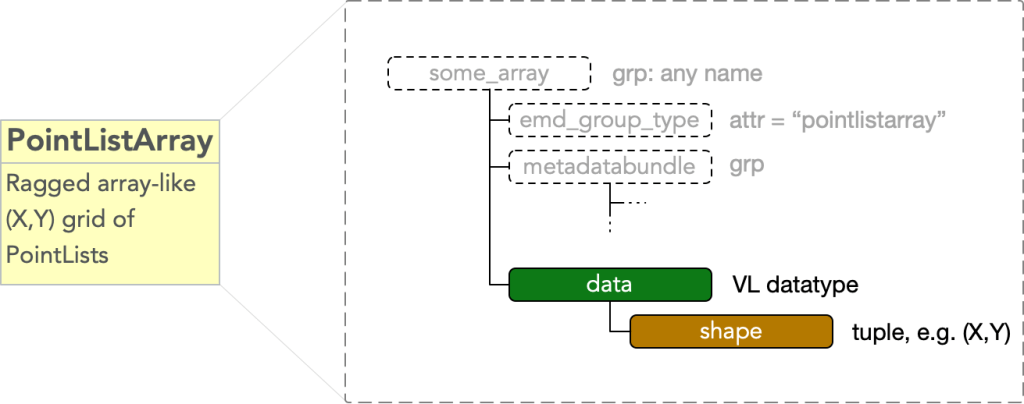
In HDF5 data structured in this way are representable as variable length datatypes. A pointlistarray must have
- attr: “emd_group_type” = “pointlistarray”
and a dataset
- dset: “data”
which is of a variable length datatype reflecting the M fields encoding the data embedding space and an
- attr: “shape”
equal to a length N tuple of integers describing the grid shape.
For example, using h5py to interface between Python and HDF5, if group is an h5py.Group instance then the Python code
dataset = group.create_dataset(
"some_name",
shape = (8,8),
dtype = h5py.special_dtype( vlen = 'uint16' )
)makes an (8, 8) shaped 2+1 dimensional ragged array of 16-bit unsigned integers. Alternatively, the code
dataset = group.create_dataset(
"some_name",
shape = (8,8),
dtype = h5py.special_dtype(
vlen = [('x','<f8'),('y','<f8'),('z','<f8')]
)
)also makes an (8, 8) shaped 2+1 dimensional ragged array, this time with the data points embedded in a 3D space (‘x’, ‘y’, ‘z’) and with each of these coordinates specified by a 64-bit float.
Group Type: Custom
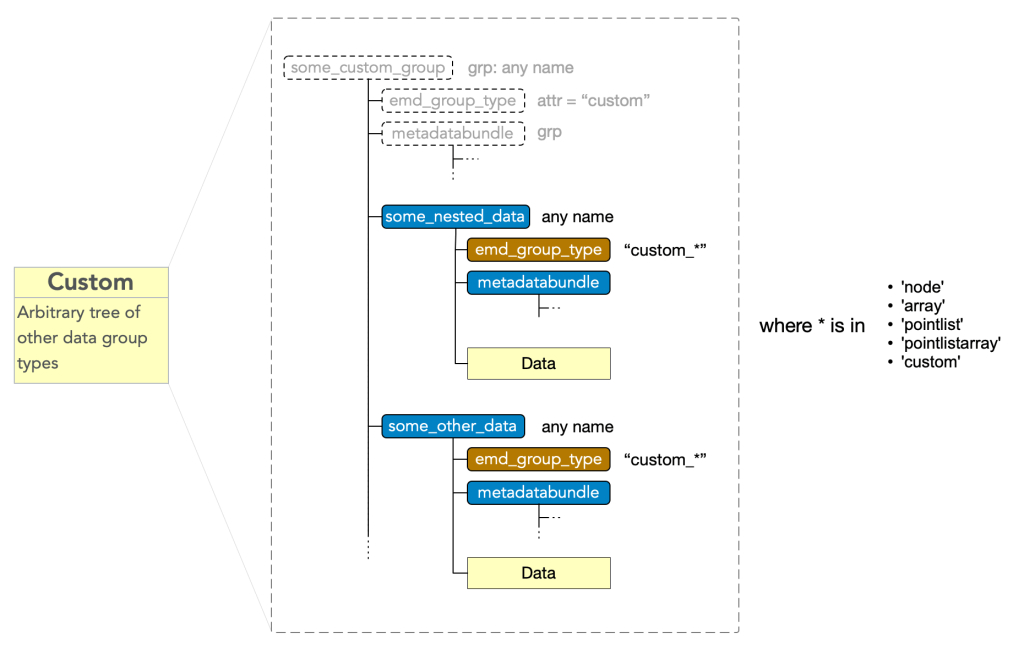
Custom type nodes enables composition data group types, encompassing a collection or nested collection of data blocks into a single object. A custom group must have
- attr: “emd_group_type” = “custom”
and may have any number of subgroups which have an
- attr: “emd_group_type” = “custom_*”
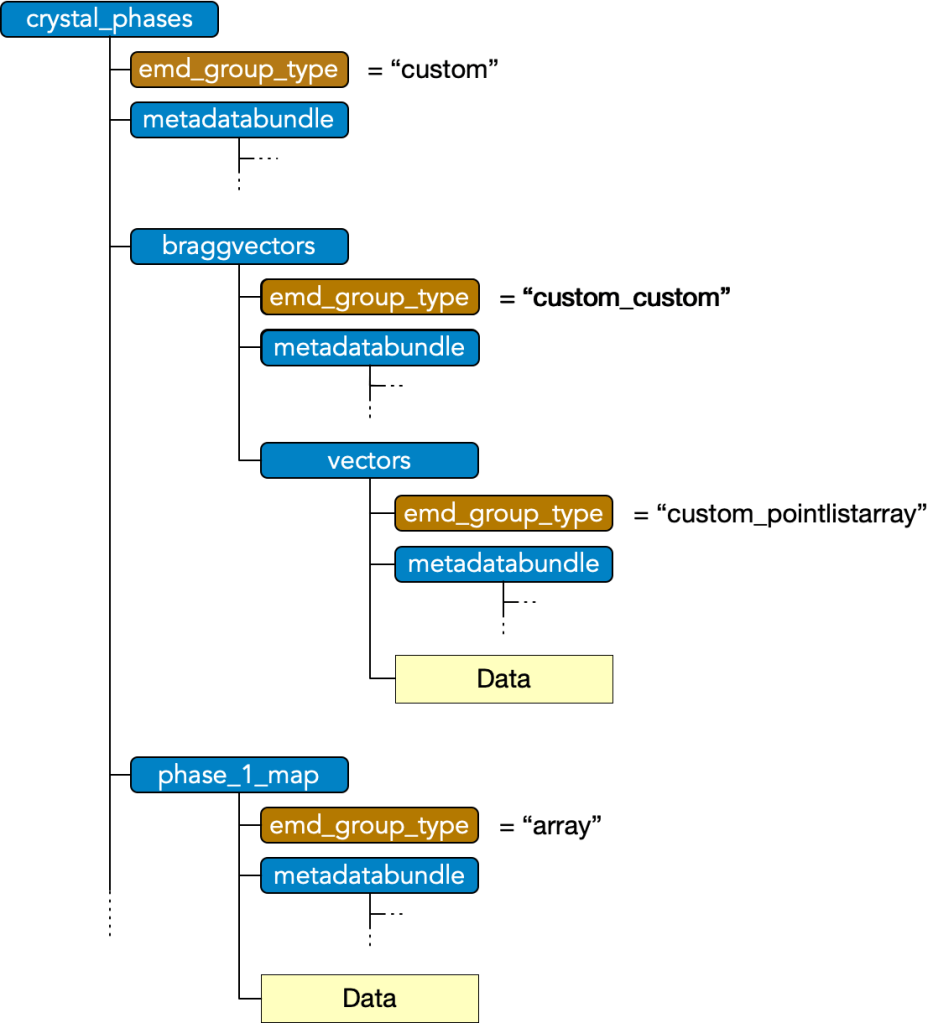
where * is a valid data type excluding the root type, i.e. must be in (“node”, “array”, “pointlist”, “pointlistarray”, “custom”). These subgroups contain the data and metadata of the corresponding EMD group types, however, are not considered tree nodes themselves beyond their role as components of the top-level custom group’s data block.
Note that “custom_custom” is possible, permitting nesting, and that in nestings beyond the first, the leading “custom_” should still only appear once, i.e. “custom_custom_*” is not valid.
Metadata
Every node may contain a group called “metadatabundle” containing any number of metadata groups.
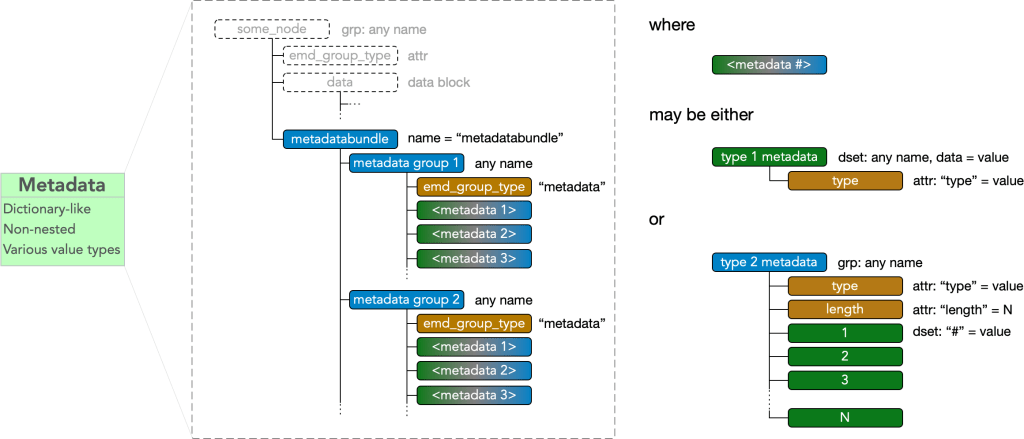
Metadata groups must contain an
- attr: “emd_group_type” = “metadata”
and contains any number of metadata items. A metadata item may be of type I, type II, or type III.
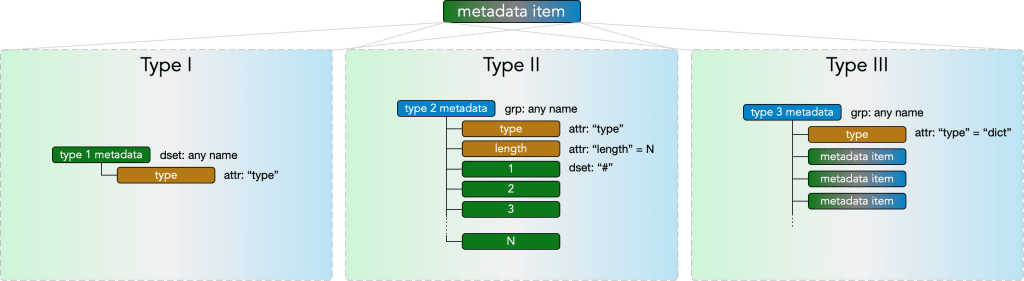
Type I metadata
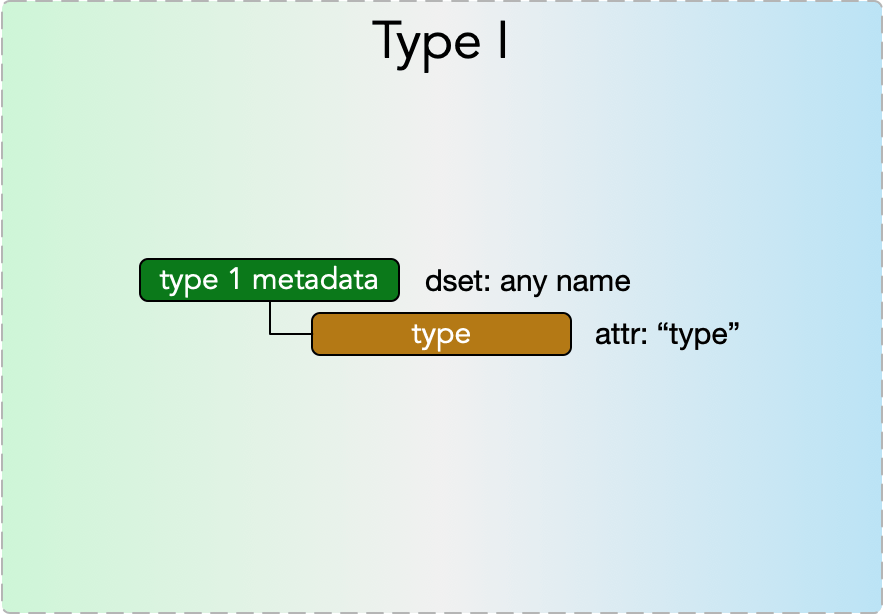
Type I metadata items represent a single value. They consist of a
- dset: any name
with an
- attr: “type”
where the permissible values of the dataset depend on the value of the “type” attribute, according to the Type I Metadata Table below.
| “type” | dtype of dataset value |
| “number” | number |
| “bool” | boolean |
| “string” | string |
| “array” | numpy ndarray |
| “None” | “_None”* |
| “tuple” | tuple of numbers |
| “list” | list of numbers |
(*) indicates a bytestring, i.e., for attr: “type” = “None” the dataset’s value is the bytestring “_None”.
Type II Metadata
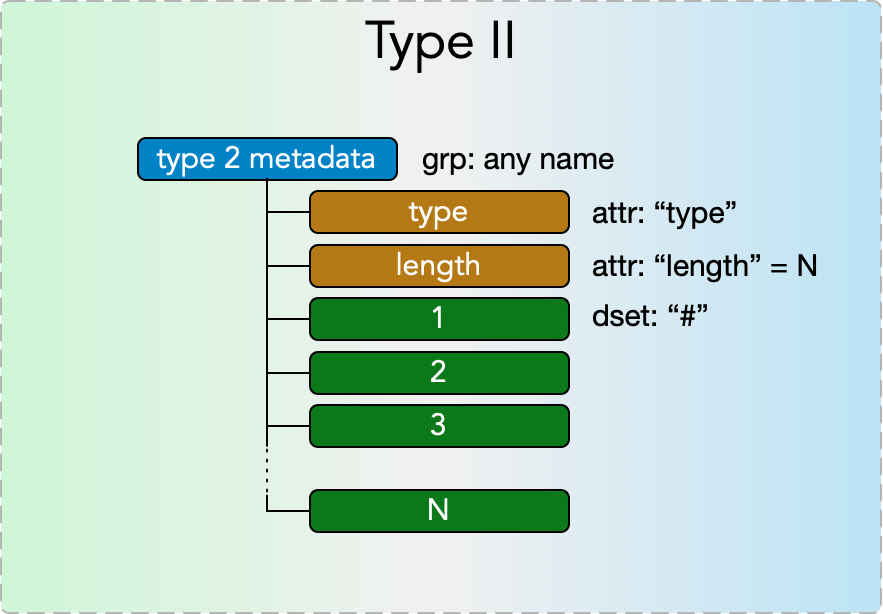
Type II metadata items represent singly nested collections of metadata of a single type, e.g. a list of arrays. They consist of a
- grp: any name
with
- attr: “type”
- attr: “length”
where the type attribute must be one of the strings from the Type II Metadata Table below and the length attribute is a nonnegative integer N. The group further contains N datasets
- dset: “#”
for each # in (1…N), with each dataset’s content’s datatype specified by the type attribute according to the Type II Metadata Table. Note that in a given type II metadata item all the datasets must have values of the same datatype.
| “type” | dtype of dataset values |
| “tuple_of_tuples” | tuples |
| “tuple_of_arrays” | numpy ndarrays |
| “tuple_of_strings” | strings |
| “list_of_arrays” | numpy ndarrays |
| “list_of_strings” | strings |
Type III Metadata
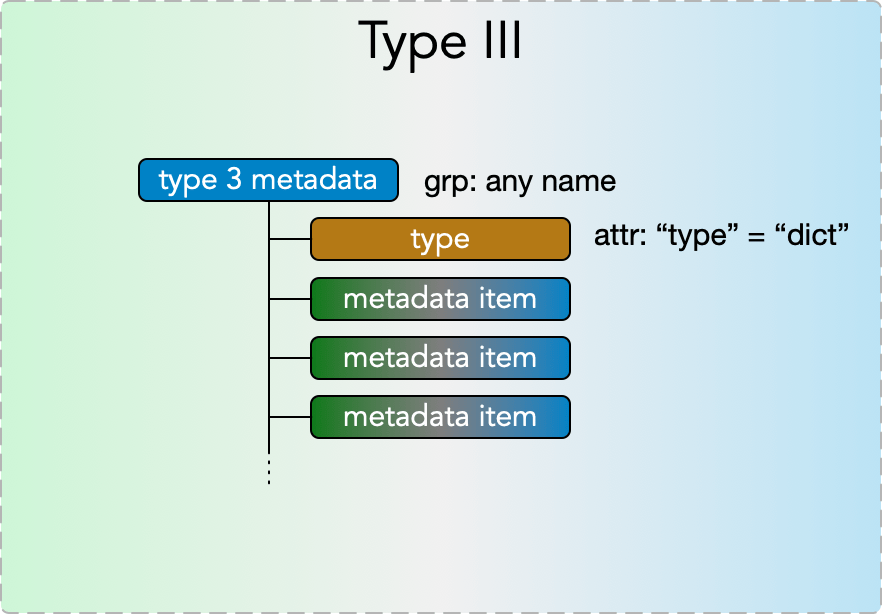
Type III metadata items are containers of any number of type I, type II, and type III items, enabling nested metadata trees of any depth. They consist of a
- grp: any name
with
- attr: “type” = “dict”
and with any number of type I, II, and III metadata items placed in the group.
Reading and writing EMD 1.0
The emdfile Python package interfaces between Python and EMD 1.0 formatted HDF5 files. It can read EMD 1.0 as well as EMD 0.1 files. Additionally, it’s designed to simplify saving and retrieving data in any Python analysis code. emdfile is available here.
For reading, any HDF5 reader should also work. The HDF5 group provides a free HDF5 viewing client called HDFView. In Python, HDF5 files can be traversed and read using the h5py package.
For writing, any file written to the specifications above in any lower level file format may be considered a valid EMD 1.0 file. If you’re writing or interested in writing data using the EMD 1.0 standard, please let us know!
