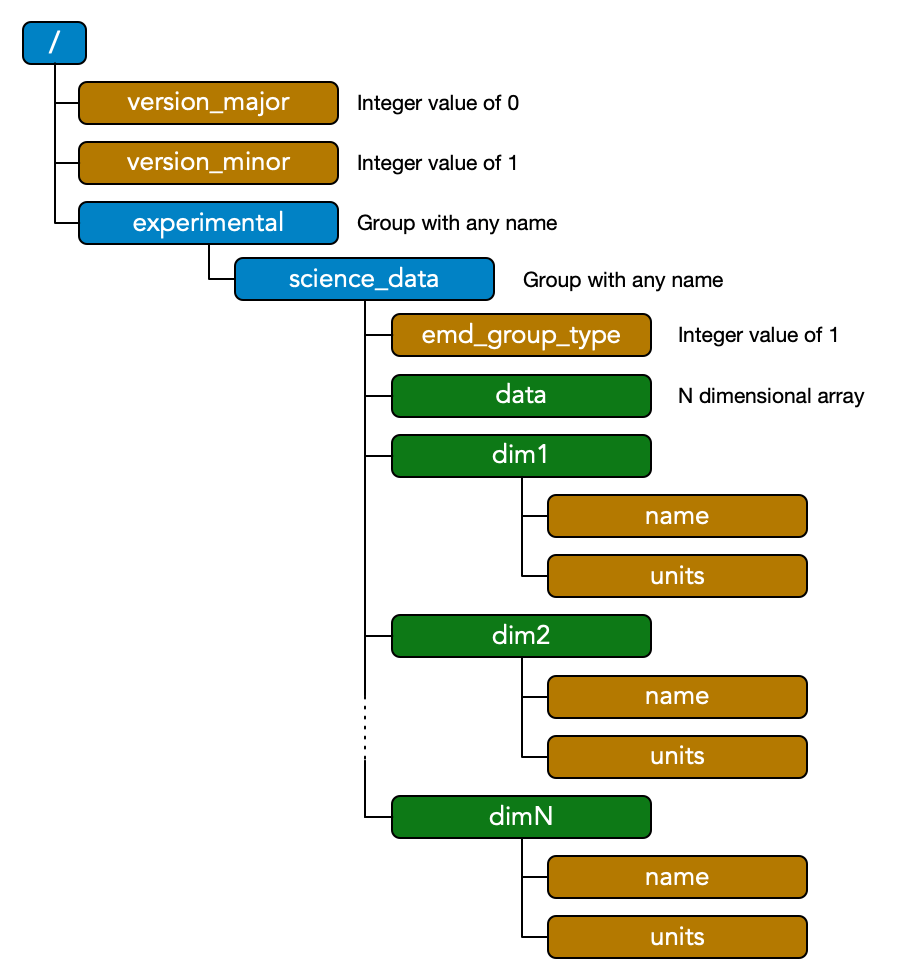
EMD v0.1 defines a specification for storing any number of N-dimensional arrays, each with a set of N 1D vectors which calibrate its axes. EMD 0.1 files must be valid HDF5 files. Below, colors and diagrams refer to HDF5 objects (groups, datasets and attributes) as discussed here.
Each N-dimensional dataset in an EMD 0.1 file is stored in an EMD data group, which is marked as such with an attribute named emd_group_type that has an integer value of 1. There are no restrictions on the name of the group. Any number of EMD data groups may be placed anywhere in the HDF5 file. However we do not recommend placing EMD data groups inside the root folder of an EMD file; rather they should be placed inside a group with a descriptive name.
EMD 0.1 Specification
Each valid EMD data group contains multiple HDF5 datasets. The first is the data itself, corresponding to a dataset named “data” and containing an N dimensional array. Additionally, the EMD data groups contains N datasets named “dim#“, where # ranges from 1 to N. The values of these datasets correspond to the coordinates along their corresponding dimension. For example the x dimension of a 1024^2 pixel micrograph with a pixel size of 0.02 nm would have dim1 values of [0, 0.02, 0.04, … 20.46]. The dim# datasets should each contain two attributes, name and units, which should be UTF-8 encoded strings, and correspond to the name of the dimension calibrated by this vector and the units it is specified in, respectively. For example name = “x” and units = “[n_m]” are common values for dim1. The data group may also optionally include a “name” and “units” attribute, however, they are not required.
If the dim# vectors have a constant step size (i.e. linearly spaced coordinates), you may remove all entries except for the first two values. To be consistent with the above description, these values will therefore be equal to [offset offset+step].
The only other requirement of the EMD specification is to attach two attributes, “version_major” and“version_minor”, to the root group. The version attributes allows a processing routine to check if an HDF5 file is a valid EMD 0.1 file. A minimal EMD file for this specification with a single N-dimensional dataset therefore looks like the diagram drawn above.
Metadata
One benefit of HDF5 is its ability to include arbitrary amounts of metadata of arbitrary types. Large arrays of values should be stored in datasets, and single values or small arrays can be stored in attributes. We do not place any restrictions on including additional data in EMD files. Extra datasets and attributes can even be freely added to EMD data groups at any level.
However, some recommendations on how to store experimental data are included here. These guidelines are intended to make it easier for different applications that use the EMD format to communicate necessary values, such as microscope parameters. Our recommendations span two areas: naming conventions, and structure.
Naming conventions
We recommend all group and attribute names be lowercase and single-word, if possible. If multiple word names are needed, words are separated by the “_” underscore character. Units are specified inside repeated square brackets with their SI prefixes placed in front of the unit itself and separated by an underscore. Raising a unit to a power other than 1 is specified with the “^” caret character, which includes inverse units prefaced by a negative sign, i.e. “^-1”. Adopting these guidelines make the units easily machine readable. Some examples include:
| Unit Type | Unit Name | EMD Unit |
| Length | nanometers | [n_m] |
| Electric potential | kilovolts | [k_V] |
| Electron interaction parameter | radians per nanometer squared | [rad][n_m^-2] |
| Force | Newtons | [N] or [k_g][m][s^-2] |
Metadata Structure
Metadata in the EMD format is typically divided into various top-level groups. Each group can contain any number of associated attributes, as well as additional subgroups. The currently recommended top-level metadata groups are:
A. Microscope
This group contains all of the relevant microscope (and therefore imaging) settings. These include parameters such as accelerating voltage, pixel size / magnification, beam current, and many others. Long lists of closely related parameters can be further lumped into subgroups. For example, the hardware corrector of an aberration-corrected TEM records many aberration parameters. Each of the (many) aberration parameters it can measure has a magnitude, angle and unit. Rather than clutter up the microscope group, we recommend placing these parameters in a subgroup aberrations.
B. Sample
We have found it very useful to include metadata tags related to the sample being imaged. These tags could include the material being imaged, sample processing information, or even qualitative comments related to the sample quality. Recording this information during the experiment greatly aids quality assessments at a future date.
C. User
All information related to the microscope operator (or the person who simulated the data) can be stored here. Information such as the user’s name, institution, department and contact email are very useful for tracing the provenance of experimental data. It also aids in database applications for large volumes of microscopy data.
D. Comments
This is a special group that contains time-stamped information on the EMD dataset. In particular we recommend that whenever the data is modified, this information be appended to the comments group. This will prevent potential mixups between raw and filtered data.
Each of these groups are optional. However using standard conventions for metadata information will make it much easier to develop software and algorithms that make use of the EMD format.
